This page contains links to bioinformatics resources useful to track the evolution and progression of the SARS-CoV-2 virus as well as to manage genomics data of the virus.
Portals with COVID-19 genomics and epidemiologics information
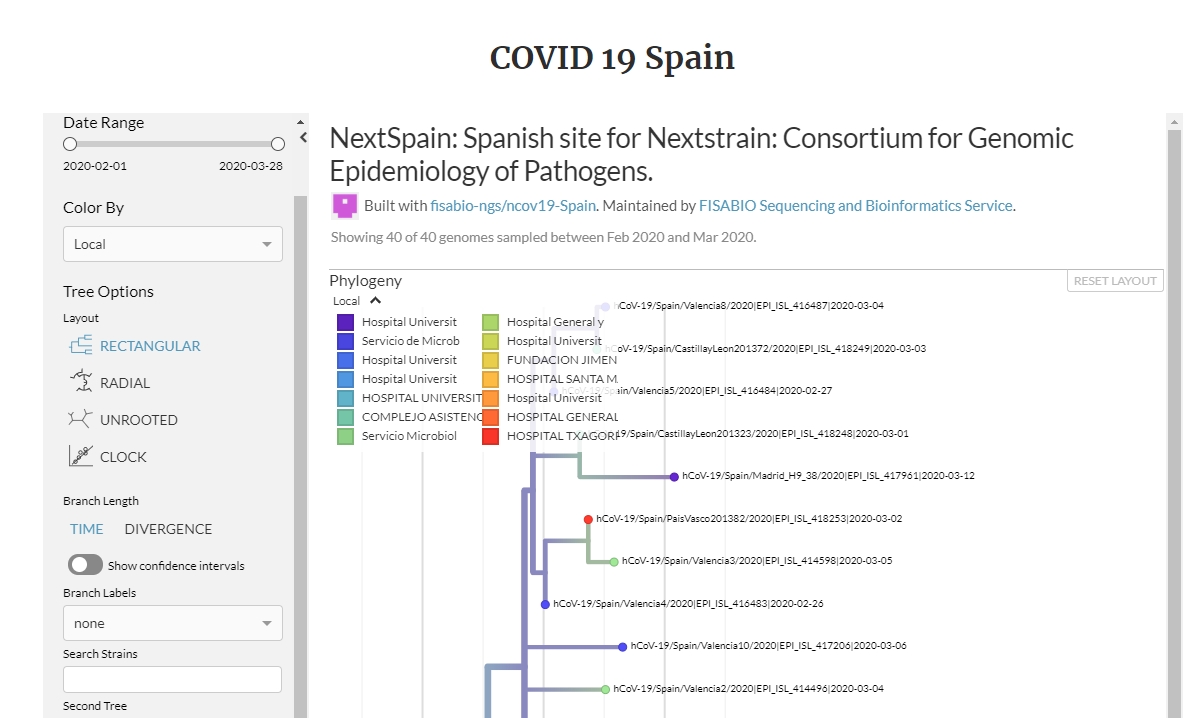
NEXTSTRAIN
Nextstrain is an open-source project to harness the scientific and public health potential of pathogen genome data. They provide a continually-updated view of publicly available data with powerful analytics and visualizations showing pathogen evolution and epidemic spread.
NCBI Virus
NCBI Virus is a community portal for viral sequence data from RefSeq, GenBank and other NCBI repositories. To find, retrieve and analyze data, please select an option below.
GISAID
The GISAID Initiative promotes the international sharing of all influenza virus sequences, related clinical and epidemiological data associated with human viruses, and geographical as well as species-specific data associated with avian and other animal viruses, to help researchers understand how the viruses evolve, spread and potentially become pandemics.
COVID-19 Disease Map
They share resources and best practices to develop a COVID-19 disease map. Aiming to set up a map of mechanisms focused on host-pathogen interactions, specific to the SARS-CoV-2 virus.
EBI - COVID-19 Portal
The COVID-19 Portal will enable researchers to upload, access and analyse COVID-19 related reference data and specialist datasets.
In the first instance, as the COVID-19 Portal is being built, all COVID-19 data stored at EMBL-EBI is being aggregated into a dedicated page on the Pathogen Portal. EMBL-EBI expects that during April, a first version of the COVID-19 Portal will be live. Watch this page for updates.
In the first instance, as the COVID-19 Portal is being built, all COVID-19 data stored at EMBL-EBI is being aggregated into a dedicated page on the Pathogen Portal. EMBL-EBI expects that during April, a first version of the COVID-19 Portal will be live. Watch this page for updates.
CoV2ID: Detection and Therapeutics Oligo Database for SARS-CoV-2
The CoV2ID evaluates molecular methods for detection of SARS-CoV-2 and treatment of COVID-19. The database evaluates the available oligonucleotide sequences (PCR primers, RT-qPCR probes, etc.) considering the genetic diversity of the virus. Updated sequences alignments are used to constantly verify the theoretical efficiency of available testing methods. Detailed information on available detection protocols are also available to help laboratories implementing SARS-CoV-2 testing.
Bioinformatics resources
CoV-Hipathia
COVID19 PATHWAY INTERPRETATION AND ANALYSIS
This web tool implements a mechanistic model of human signaling for the interpretation of the consequences of the combined changes of gene expression levels and/or genomic mutations in the context of signalling pathways.
This web tool implements a mechanistic model of human signaling for the interpretation of the consequences of the combined changes of gene expression levels and/or genomic mutations in the context of signalling pathways.
Artic
The ARTIC network is making available a set of materials to assist groups in sequencing the virus including a set of primers, laboratory protocols, bioinformatics tutorials and datasets, mainly focused around the use of the portable Oxford Nanopore MinION sequencer.
SARS-CoV-2
SARS-CoV-2 Initial analysis of COVID-19 data using Galaxy, BioConda and public research infrastructures
This repo serves as a companion to our study describing the analysis of early COVID-19 data. We currently feature two different projects: one analyzing the COVID-19 Genome and the other performing virtual screening on the SARS-CoV-2 main protease.
It contains descriptions of workflows and exact versions of all software used. The goals of this study were to:
- Underscore the importance of access to raw data
- Demonstrate that existing community efforts in curation and deployment of biomedical software can reliably support rapid reproducible research during global crises
BU-ISCIII / SARS-Cov2_analysis
Information collected by the Bioinformatics Unit of the ISCIII
This repo will contain all the information we can collect about the bioinformatics analysis of SARS-Cov2. We will try to include:
- General information about the virus: genomics, epidemiology, molecular mechanisms,etc
- Wetlab protocols: we are no wetlab experts, here we collect another groups/bibliography approximations that are currently being used. Knowing the protocols is imperative for develop comprenhensive pipelines addecuated to the data.
- Data resources: summary of resources for covid19 (raw data, genomes, etc) and sofware/pipelines/initiatives already going on so we can all share our knowledge.
- Analysis pipelines: different analysis aproximations depending on the data: experiment (metagenomics, sispa, amplicons) and sequencing platform (Illumina, pacbio, nanopore).
Breaking-Cas
Breaking-Cas—interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes at BioinfoGP CNB, with the SARS2 genomes
Onramp Bio
Coronavirus Community Space to provide open-access of ROSALIND, a discovery and collaboration platform, for fellow scientists who wish to join a community effort to share, discover and accelerate our collective understanding of Coronavirus.
SARS-CoV-2 Sequencing Resources
Crowd-sourced collection of information, documentation, protocols and other resources for public health laboratories intending to sequence SARS-CoV-2 coronavirus samples. By Duncan MacCannell from the Office of Advanced Molecular Detection (AMD ) at the Centers for Disease Control and Prevention (CDC).
COVID-19 Pathway Collection
WikiPathways is a database of biological pathways maintained by and for the scientific community. This special subset of disease pathways is being highlighted during the current COVID-19 crisis.
Comparison of COVID-19 trajectories worldwide
BIIT is research group focusing in bioinformatics, data mining, image analysis and e-health data solutions. Their work concentrates on analysing omics data and developing novel methods and applications for easier data analysis for the wider public.
ELIXIR support for SARS-CoV-2 research
As the ESFRI Research Infrastructure for life science data, ELIXIR Nodes provide a range of services that can be used by researchers and consortia working on SARS-CoV-2 research.
Disease Maps and text mining for drug prediction
High quality knowledge combined with automated text mining and modeling can guide drug profiling research
Coronavirus Discovery resource
This is a Coronavirus research view of can SAR BLACK that we provide freely and openly to the research and drug discovery community to support them in the battle against Covid-19. This is a community resource and we invite the coronavirus research community to input (contact us)